Code
# Print "Hello, World!" to the console
print("Hello, World!")[1] "Hello, World!"Download and install R from the Comprehensive R Archive Network (CRAN) and choose the relevant OS (Windows,mac,linux).
RStudio is a recommended integrated development environment (IDE) for R. Download and install RStudio form POSIT and choose the relevant OS (Windows,mac,linux).
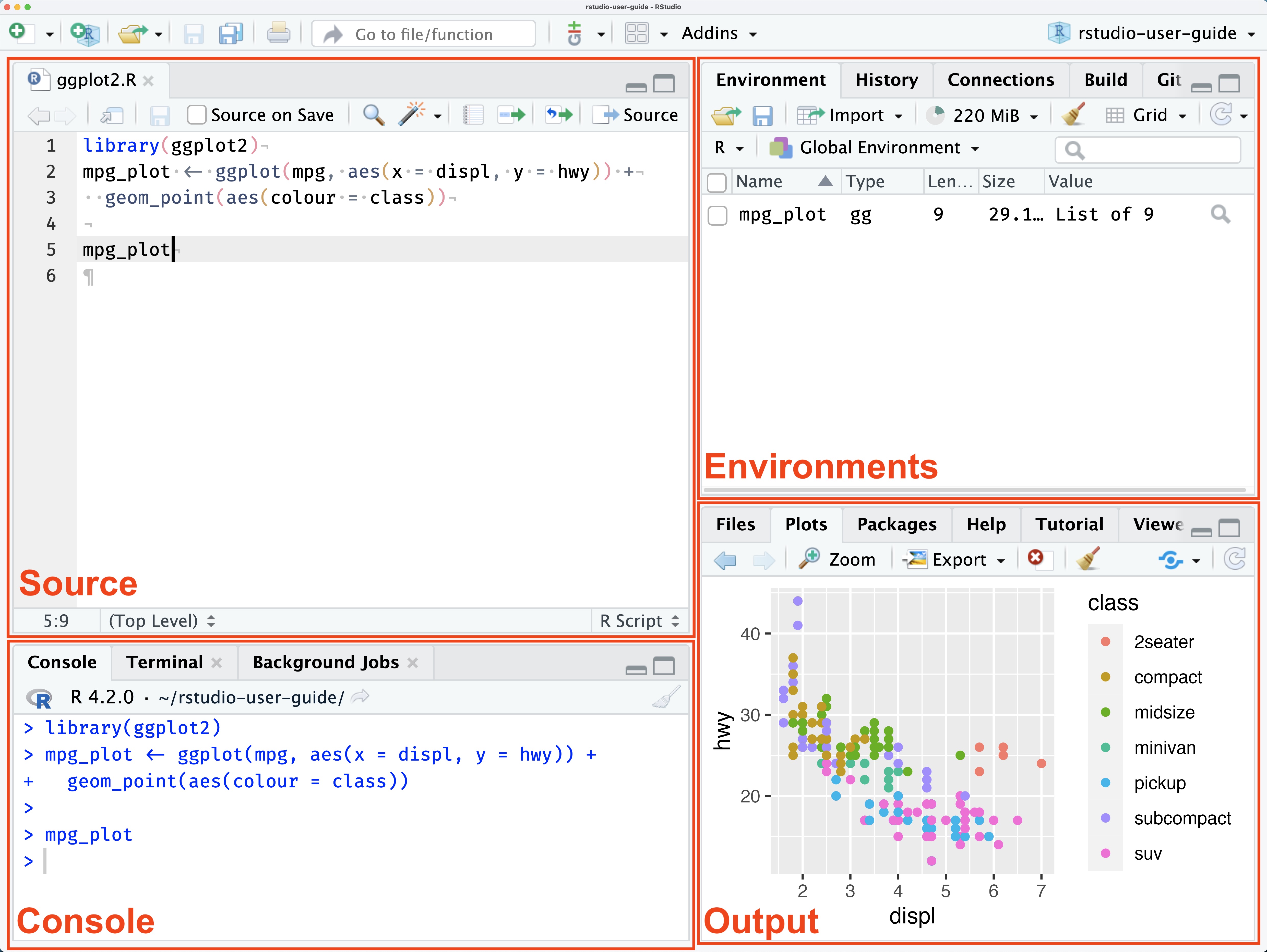
This panel is where you write and edit your R scripts and R Markdown documents.
This is where R code is executed interactively.
R is a powerful programming language used extensively for statistical computing and graphics. It provides a wide array of techniques for data analysis, including linear and nonlinear modeling, classical statistical tests, time-series analysis, classification, clustering, and more. Its syntax allows users to easily manipulate data, perform calculations, and create graphical displays. Here’s a breakdown of some fundamental aspects of R syntax and an example to illustrate how it works.
Variables: In R, you can create variables without declaring their data type. You simply assign values directly with the assignment operator <- or =.
Comments: Comments start with the # symbol. Everything to the right of the # in a line is ignored by the interpreter.
Vectors: One of the basic data types in R is the vector, which you create using the c() function. Vectors are sequences of elements of the same type.
Functions: Functions are defined using the function keyword. They can take inputs (arguments), perform actions, and return a result.
Conditional Statements: R supports the usual if-else conditional constructs.
Loops: For iterating over sequences, R provides for, while, and repeat loops.
Packages: R’s functionality is extended through packages, which are collections of functions, data, and compiled code. You can install packages using the install.packages() function and load them with library().
.R extension) without opening an interactive R session.Creating and using R scripts in RStudio is a fundamental skill for anyone working with data in R. RStudio, being a powerful IDE for R, streamlines the process of writing, running, and managing R scripts. Here’s a concise guide based on insights from various sources:
Start a New Script: To begin, navigate to File -> New File -> R Script. This opens a new script tab in the top-left pane where you can write your code.
Writing Code: You can type your R code directly into this script pane. Common tasks include importing data, data manipulation, statistical analysis, and plotting. For instance, to create and print a variable, simply type something like result <- 3 followed by print(result) to see the output in the Console pane.
Running Code: To execute your code, you can click the Run button at the top of the script pane, or use keyboard shortcuts (e.g., Ctrl+Enter on Windows or Cmnd+Enter on Mac). The output will appear in the Console pane at the bottom.
Below are a few examples of basic R scripts that demonstrate common tasks in R.
A simple script that prints “Hello, World!” to the console.
# Print "Hello, World!" to the console
print("Hello, World!")[1] "Hello, World!"This script performs basic arithmetic operations and prints the results.
# Perform arithmetic operations
add <- 5 + 3
# Print the results
add[1] 8Data types refer to the kind of data that can be stored and manipulated within a program. In R, the basic data types include:
Use <- or = for assigning values, e.g., x <- 10 or x= 10
x=5
x[1] 5y <- 3
y[1] 3Use # for comments, e.g., # This is a comment.
# symbol, is considered as comment.# Assigning values to x
x=5
x[1] 5+, -, *, /, ^
# Perform arithmetic operations
add <- 5 + 3
# Print the results
add[1] 8Division (/) operator - Divides the first number or vector by the second, element-wise.
x=5
y=3
x/y[1] 1.666667Square (^) operator - Squares the first number by the second.
x=5
x^2[1] 25Includes ==, !=, >, <, >=, <=.
Equality: == checks if two values are equal.
# Equality 5 == 3
x <- 5
y <- 3
x == y [1] FALSEInequality: != checks if two values are not equal.
# Inequality 5 != 3
x != y [1] TRUEGreater than: > checks if the value on the left is greater than the value on the right.
# Greater than 5 > 3
x > y [1] TRUELess than: < checks if the value on the left is less than the value on the right.
# Less than 5 < 3
x < y [1] FALSEGreater than or equal to: >= checks if the value on the left is greater than or equal to the value on the right.
# Greater than or equal to 5 >= 3
x >= y [1] TRUELess than or equal to: <= checks if the value on the left is less than or equal to the value on the right.
# Less than or equal to 5 <= 3
y <= x [1] TRUEc() function combines values into a vector. It’s the most common method for creating vectors., , 1
[,1] [,2] [,3]
[1,] 1 3 5
[2,] 2 4 6
, , 2
[,1] [,2] [,3]
[1,] 7 9 11
[2,] 8 10 12
, , 3
[,1] [,2] [,3]
[1,] 13 15 17
[2,] 14 16 18
, , 4
[,1] [,2] [,3]
[1,] 19 21 23
[2,] 20 22 24Consists inbuilt functions like sum(), length(), sqrt(),mean(), summary(), View()
The sum() function calculates the total sum of all the elements in a numeric vector.
The length() function returns the number of elements in a vector (or other objects).
The sqrt() function calculates the square root of each element in a numeric vector.
The mean() function calculates the arithmetic mean (average) of the elements in a numeric vector.
The summary() function in R provides a concise statistical summary of objects like vectors, matrices, data frames, and results of model fitting.
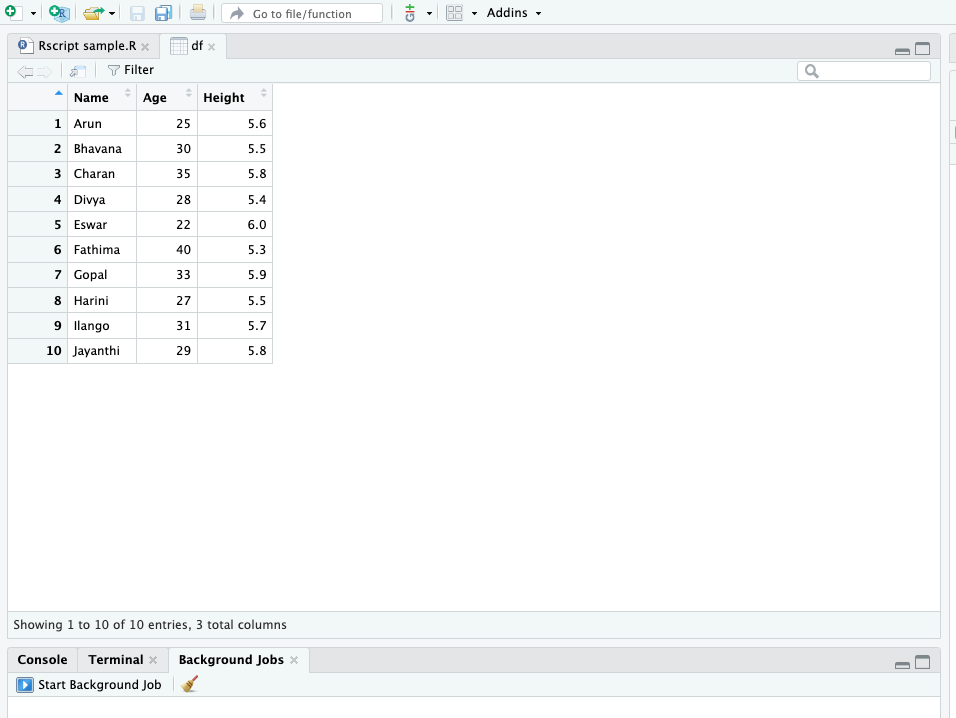
data.frame() function is used to create data frames, which are table-like structures consisting of rows and columns. - Data frames are one of the most important data structures in R, especially for statistical modeling and data analysis.
creating data frame
# Creating a simple data frame
students <- data.frame(
Name = c("Arun", "Bhavana", "Charan", "Divya", "Eswar",
"Fathima", "Gopal", "Harini", "Ilango", "Jayanthi"),
Age = c(25, 30, 35, 28, 22, 40, 33, 27, 31, 29),
Height = c(5.6, 5.5, 5.8, 5.4, 6.0, 5.3, 5.9, 5.5, 5.7, 5.8)
)
students Name Age Height
1 Arun 25 5.6
2 Bhavana 30 5.5
3 Charan 35 5.8
4 Divya 28 5.4
5 Eswar 22 6.0
6 Fathima 40 5.3
7 Gopal 33 5.9
8 Harini 27 5.5
9 Ilango 31 5.7
10 Jayanthi 29 5.8The head() function in R is used to display the first few rows of a dataset, making it a useful tool for quickly inspecting large data frames or matrices.
Name Age Height
1 Arun 25 5.6
2 Bhavana 30 5.5
3 Charan 35 5.8
4 Divya 28 5.4
5 Eswar 22 6.0
6 Fathima 40 5.3View() function is used to invoke a spreadsheet-like data viewer on a data frame, matrix, or other objects that can be coerced into a data frame. - This function is particularly useful during interactive sessions to inspect data visually.
Use for, while.
The for loop in R is used to iterate over a sequence (like a vector or a list) and execute a block of code for each element in the sequence.
[1] 1
[1] 2
[1] 3
[1] 4
[1] 5The while loop executes a block of code as long as the specified condition is TRUE
while loop
# Print numbers from 1 to 5 using while
i <- 1 # Initialize counter
while(i <= 4) {
print(i)
i <- i + 1 # Increment counte
}[1] 1
[1] 2
[1] 3
[1] 4In RStudio, you can create a new R Markdown file via the menu: File -> New File -> R Markdown....
 This opens a dialog where you can choose the output format and other options.
This opens a dialog where you can choose the output format and other options.
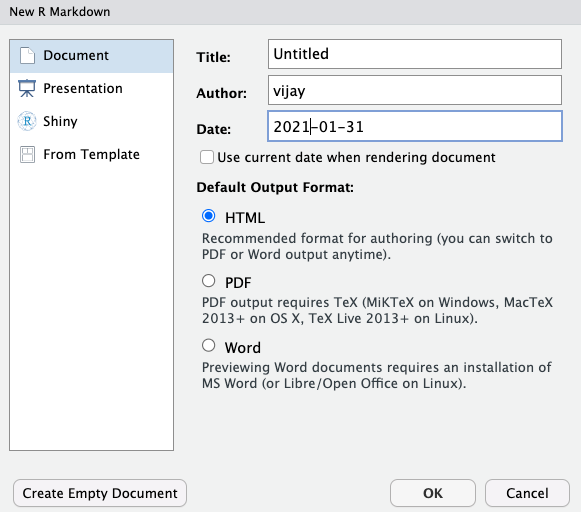 Enter a title, author and date, check html for html output and click ok.
Enter a title, author and date, check html for html output and click ok.
New R Markdown will open in a new window as shown below.
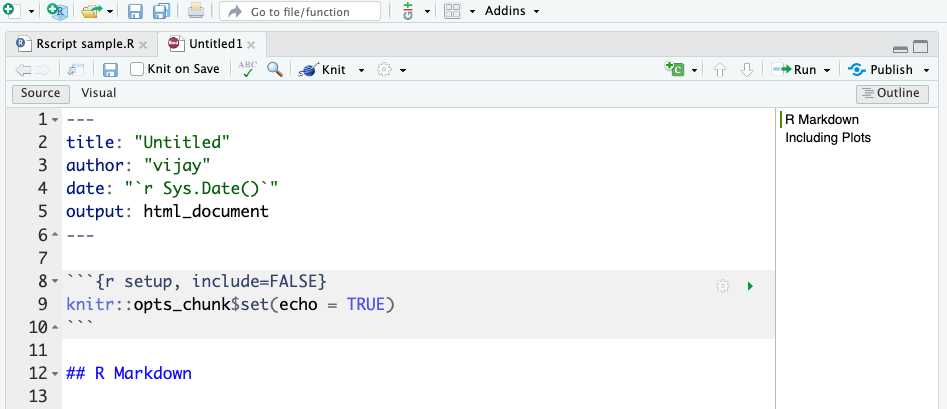
When you create a new R Markdown file in RStudio, a default template is generated with the following components:
At the top of the document, you will see a YAML metadata block, enclosed within triple dashes ---. This section specifies document settings such as the title, author, date, and output format.---
title: “Untitled”
author: “vijay”
date: “2025-04-18”
output: html_document---
Immediately following the YAML header, a setup chunk is included:
{r setup, include=FALSE}
knitr::opts_chunk$set(echo = TRUE)
# R Markdown
This is a section heading that introduces the user to R Markdown. It is placed there to provide a structured template for writing content.
options(repos = c(CRAN = "https://cran.rstudio.com/"))
knitr::opts_chunk$set(message = FALSE)Including this code at the beginning of an RMarkdown file ensures that R uses a specific CRAN repository (https://cran.rstudio.com/) for package installation and suppresses unnecessary messages in code chunks, making the document cleaner and more readable.
It should look like the below.
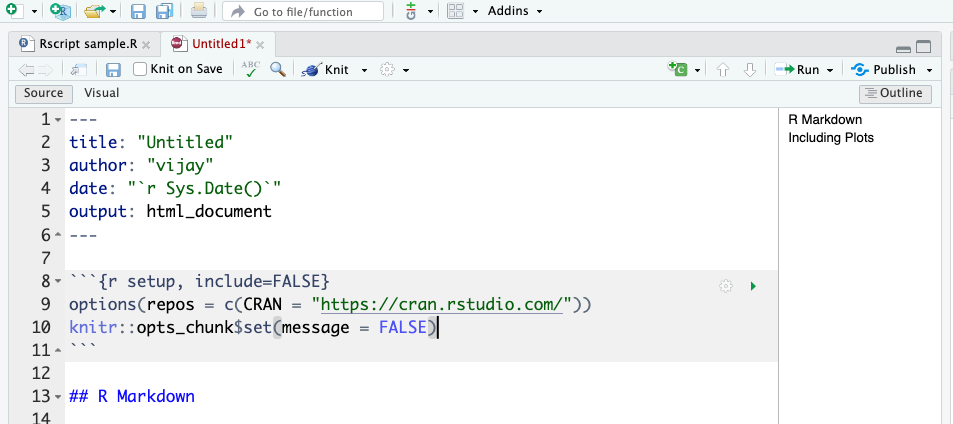
After saving the file, compile the document into your desired output format. click the Knit button in RStudio. This will execute all R code within the document and render it to the specified output format.
R Markdown supports standard Markdown syntax. Here are some basic examples:
# for headers. E.g., # Header 1, ## Header 2.**bold** for bold text and *italic* for italic text.- or * for unordered lists and numbers for ordered lists.[link text](URL) to create hyperlinks. to insert images.You can embed R code within your document by using the following syntax:
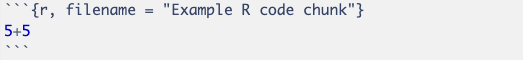
The three backticks (```) mark the beginning of a code chunk, while {r} specifies that the chunk contains R code. To properly close the code chunk, another set of three backticks (```) must be included at the end.
The code inside this chunk is executed, and its results are included in the document below the chunk.
To compile the document into your desired output format, click the Knit button in RStudio. This will execute all R code within the document and render it to the specified output format.
Dynamic Report Generation
Multiple Output Formats
Reproducibility
Ease of Use
Here’s a simple example of an R Markdown document that performs a basic data analysis:
Min. 1st Qu. Median Mean 3rd Qu. Max.
4.0 12.0 15.0 15.4 19.0 25.0 Let’s calculate summary statistics for the pressure dataset:
temperature pressure
Min. : 0 Min. : 0.0002
1st Qu.: 90 1st Qu.: 0.1800
Median :180 Median : 8.8000
Mean :180 Mean :124.3367
3rd Qu.:270 3rd Qu.:126.5000
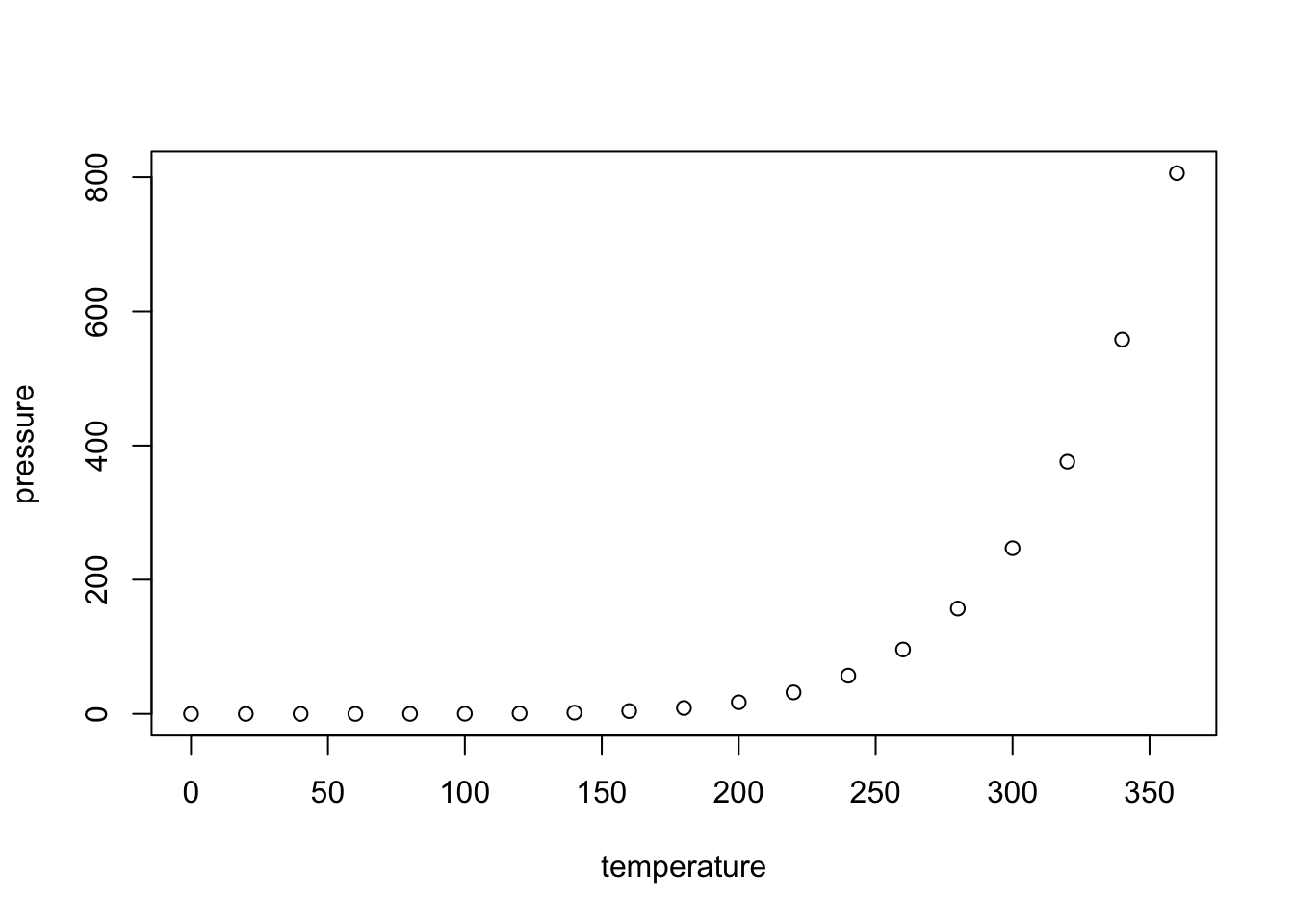
Max. :360 Max. :806.0000 You can also embed plots. For example, here’s a plot of pressure vs temperature:
The most important set of options controls if your code block is executed and what results are inserted in the finished report:
eval = FALSE prevents code from being evaluated. (And obviously if the code is not run, no results will be generated). This is useful for displaying example code, or for disabling a large block of code without commenting each line.
include = FALSE runs the code, but doesn’t show the code or results in the final document. Use this for setup code that you don’t want cluttering your report.
echo = FALSE prevents code, but not the results from appearing in the finished file. Use this when writing reports aimed at people who don’t want to see the underlying R code.
message = FALSE or warning = FALSE prevents messages or warnings from appearing in the finished file.
error = TRUE causes the render to continue even if code returns an error.
| Option | Run code | Show code | Output | Plots | Messages | Warnings |
|---|---|---|---|---|---|---|
eval = FALSE |
||||||
include = FALSE |
✓ | |||||
echo = FALSE |
✓ | ✓ | ✓ | ✓ | ✓ | |
results = "hide" |
✓ | ✓ | ✓ | ✓ | ✓ | |
fig.show = "hide" |
✓ | ✓ | ✓ | ✓ | ✓ | |
message = FALSE |
✓ | ✓ | ✓ | ✓ | ✓ | |
warning = FALSE |
✓ | ✓ | ✓ | ✓ | ✓ |
A directory in R is a folder in the computer where files are stored and accessed. R allows users to interact with directories to read data files, save outputs, and manage projects efficiently. Understanding how to check and change the working directory is crucial when dealing with file operations in R.
The working directory is the default location where R reads and writes files. When working with data files such as CSV, Excel, or text files, R looks for these files in the current working directory unless a full file path is specified.
The setwd() function allows users to change the working directory. This is useful when dealing with files located in different folders.
setwd("/Users/vijay/Library/")After setting a new directory, you can verify it by running getwd().
The getwd() function in R is used to check the current working directory. This helps users confirm where R is looking for files and where outputs will be saved.
# Get the current working directory
getwd()[1] "/Users/vijay/Library/CloudStorage/OneDrive-Personal/Documents/1 Professional/3 My Books/BAAGR"[1] "/Users/vijay/Library/"To check the available files in the current working directory, use list.files():
# List all files in the current working directory
list.files()This is helpful when verifying whether the required files exist before attempting to read them.
getwd() before reading or saving files.setwd() cautiously to avoid breaking file paths when sharing scripts across different systems.R provides a vast ecosystem of packages designed for data analysis, visualization, and statistical modeling. In this section, we will explore five essential packages:
stats: Statistical analysis and hypothesis testingplotly: Interactive visualizationstidyverse: A collection of packages for data manipulation and visualizationEach of these packages serves a crucial role in handling data efficiently and performing complex analyses in R.
The stats package comes pre-installed with R and provides essential statistical functions for data analysis.
# Descriptive statistics
summary(mtcars$mpg) Min. 1st Qu. Median Mean 3rd Qu. Max.
10.40 15.43 19.20 20.09 22.80 33.90 # t-test example
t.test(mtcars$mpg ~ mtcars$am)
Welch Two Sample t-test
data: mtcars$mpg by mtcars$am
t = -3.7671, df = 18.332, p-value = 0.001374
alternative hypothesis: true difference in means between group 0 and group 1 is not equal to 0
95 percent confidence interval:
-11.280194 -3.209684
sample estimates:
mean in group 0 mean in group 1
17.14737 24.39231
Call:
lm(formula = mpg ~ wt, data = mtcars)
Residuals:
Min 1Q Median 3Q Max
-4.5432 -2.3647 -0.1252 1.4096 6.8727
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 37.2851 1.8776 19.858 < 2e-16 ***
wt -5.3445 0.5591 -9.559 1.29e-10 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.046 on 30 degrees of freedom
Multiple R-squared: 0.7528, Adjusted R-squared: 0.7446
F-statistic: 91.38 on 1 and 30 DF, p-value: 1.294e-10The plotly package enables the creation of interactive and dynamic charts in R, making it useful for exploring data visually.
ggplot2.# Install and load the package
install.packages("plotly")An agribusiness analyst visualizes seasonal trends in crop yields using plotly, making it easier to identify patterns and variations.
The tidyverse is a collection of R packages designed for data science. It provides a structured approach to importing, manipulating, visualizing, and modeling data.
# Install tidyverse
install.packages("tidyverse")
# Load all core tidyverse packages
library(tidyverse)This loads several useful packages such as ggplot2, dplyr, tidyr, readr, purrr, tibble, stringr, and forcats.
# A tibble: 2 × 2
Name Score
<chr> <dbl>
1 John 85
2 Alice 90print(average_score)# A tibble: 1 × 1
Avg_Score
<dbl>
1 84.3# A tibble: 4 × 3
Name Subject Marks
<chr> <chr> <dbl>
1 A Math 85
2 A Science 78
3 B Math 90
4 B Science 92
library(stringr)
text <- "Welcome to R programming!"
# Convert to lowercase
lower_text <- str_to_lower(text)
# Replace "R" with "Tidyverse"
new_text <- str_replace(text, "R", "Tidyverse")
print(lower_text)[1] "welcome to r programming!"print(new_text)[1] "Welcome to Tidyverse programming!"[1] Apple Banana Cherry Banana Apple
Levels: Apple Banana Cherry